Biochemistry, Structural Biology, Molecular Biology, and Computational Chemistry
- Molecular cloning and sub-cloning, and Mutagenesis
- Expression by using yeast and E. coli systems and cell culture
- Refolding and purification of proteins by using Affinity, charge, and size exclusion chromatography.
- Charatcterization by SDS-PAGE, western blot, MALS/DLS, Mass-spectroscopy
- Biophysical characterisation using circular dichroism, fluorescence spectroscopy, small angle X-ray, scattering
- Protein ligand/drug interaction studies, enzyme kinetics, in-vitro binding assay, SPR assay
- 3-dimensional structure determination of proteins by NMR and X-ray crystallography
- Computational techniques like molecular simulation, docking and molecular modeling.
Our research interest has focused on structure-function studies of proteins using high-resolution solution NMR, circular dichroism (CD) and fluorescence spectroscopy techniques. It is amazing to see the relationship between the primary sequence of a protein to its 3-dimensional structure that defines specificity as well as diversity in its functions and how even a point mutation can sometimes disrupt or modify the functions of a protein causing in many instances abnormalities or diseases. To get a view of protein structure, function and mode of interactions at a molecular level, we use high resolution solution NMR, circular dichorism (CD), fluorescence techniques along with biochemical, molecular biology (cloning, mutagenesis) and computational chemistry.
Thus, molecular biology, biochemistry, protein chemistry, biophysical chemistry (NMR and other techniques) and computational chemistry are used in our laboratory to understand and correlate the structure-functions of both soluble and membrane bound proteins. Several projects are currently in progress in our laboratory. Research work in our laboratory is supported by funding from NSF and USDA.
Congenital Disorders of Glycosylation
We are investigating the molecular mechanisms of function of an extremely important eukaryotic membrane bound enzyme “oligosaccharyltransferase (OST)”. Oligosaccharyltransferase is a membrane associated multimeric enzyme located in the endoplasmic reticulum (ER) and is involved in co-translational N-linked glycosylation of nascent protein. Genetic defects in OST cause a series of clinical problems known as congenital disorders of glycosylation (CDG) that includes mental retardation, developmental delay, hypoglycemia, dysmorphic features, anorexia etc. Complete loss is lethal for all animals. Our group is actively involved in the structure-function studies of both yeast and human Oligosaccharyltransferase enzymes. The 3-dimensional structures of proteins are essentially “blueprints” for the development of a chemicalcompound to be a successful drug.Our group has already solved the 3-dimensional solution structure of various subunits of yeast OST enzyme. Below is 3D structure of the catalytic
C-terminal domain of yeast Stt3p.
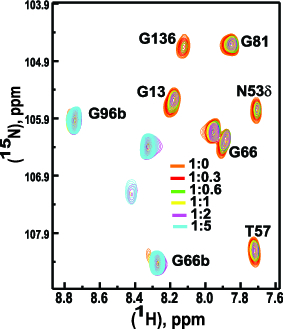
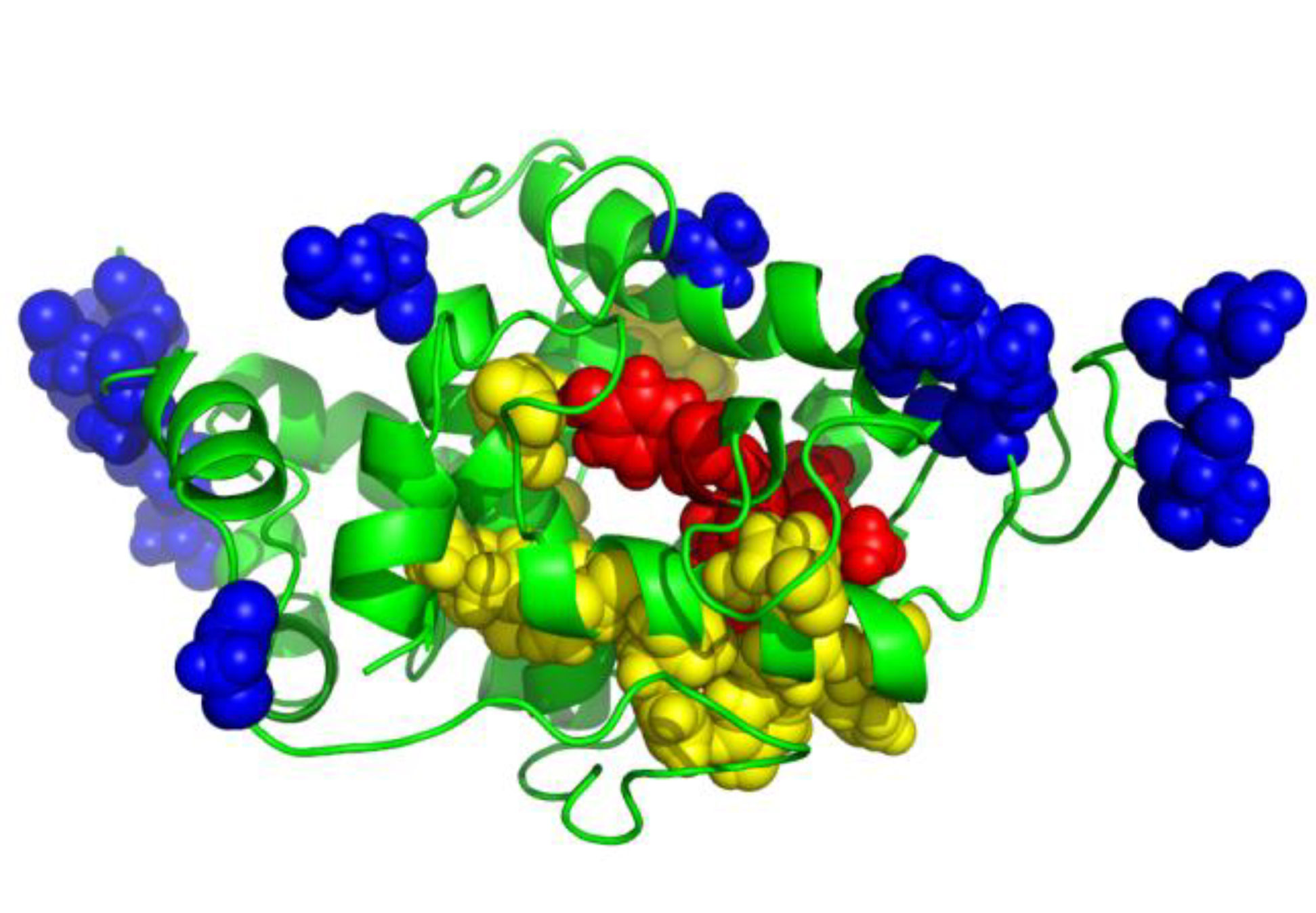 Binding site residues mapping with acceptor peptide (Hung et. al., JBC, 2012).
Binding site residues mapping with acceptor peptide (Hung et. al., JBC, 2012).
Moreover, we have cloned, successfully overexpressed, and purified both Ost4p and Stt3p subunits of yeast OST. Structural and functional characterization of Stt3p along with that of other yeast and human OST subunits is in progress in our laboratory. These studies will provide insight into the mechanisms of function of this essential and critical enzyme.
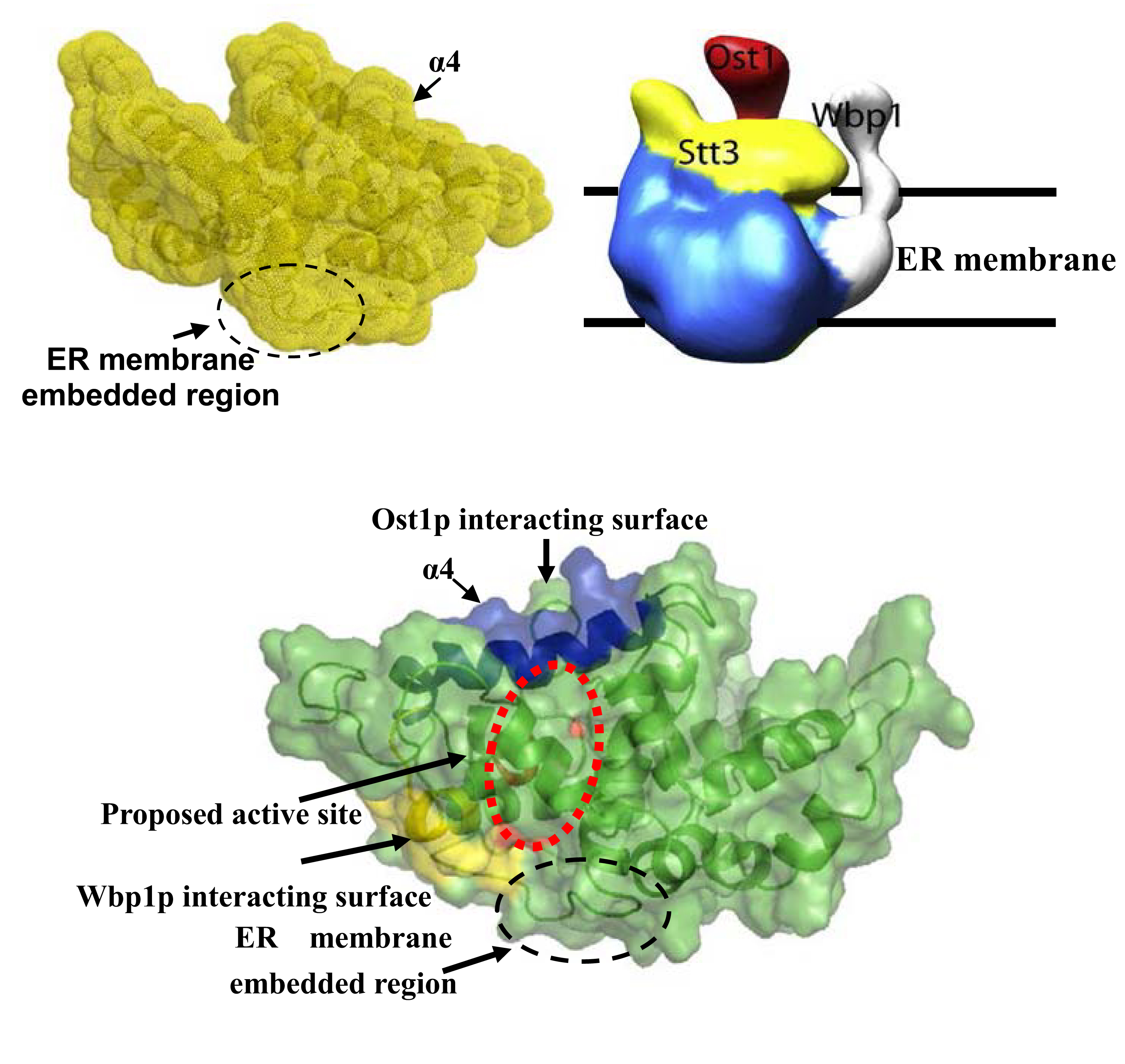
A: NMR model of catalytic domain of Stt3p, B: EM model of yeast OST complex, C: Proposed interactions with other subunits (Hung et. al., JBC, 2012).
Signal Transduction in Olfaction
We have made great progress in understanding the mechanisms of odor detection in lepidopteran moth by the olfactory receptor using the proteins of “smell” or odorant binding protein (OBP). Our solution NMR structure of an OBP revealed the details of the odorant-binding site and provided the first insight into the nature of the odor uptake mechanism at neutral pH (Mohanty et. al., JMB, 2004). Moreover, from the NMR structure of this protein at acidic pH, we reported a novel mechanism of odor release, triggered by the protonation of key histidine residues near the olfactory neuron where the membrane potential decreases the local pH (JMB, 2005; JBC, 2009 and Biochemistry, 2013). Our current research efforts are focused to test the above model of ligand release by mutational studies and to address the question of molecular recognition and mechanisms of substrate specificity by OBPs of different moth species.
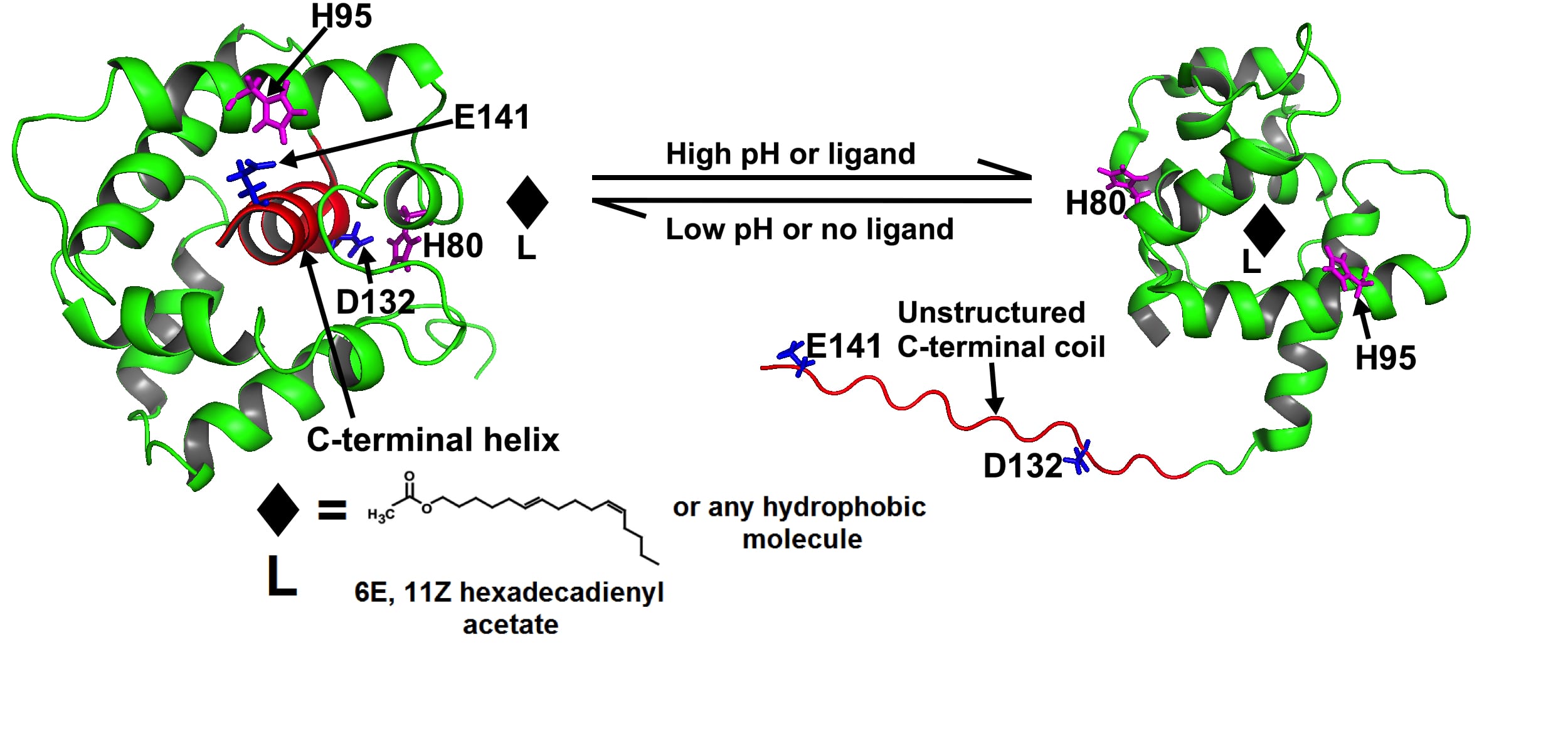
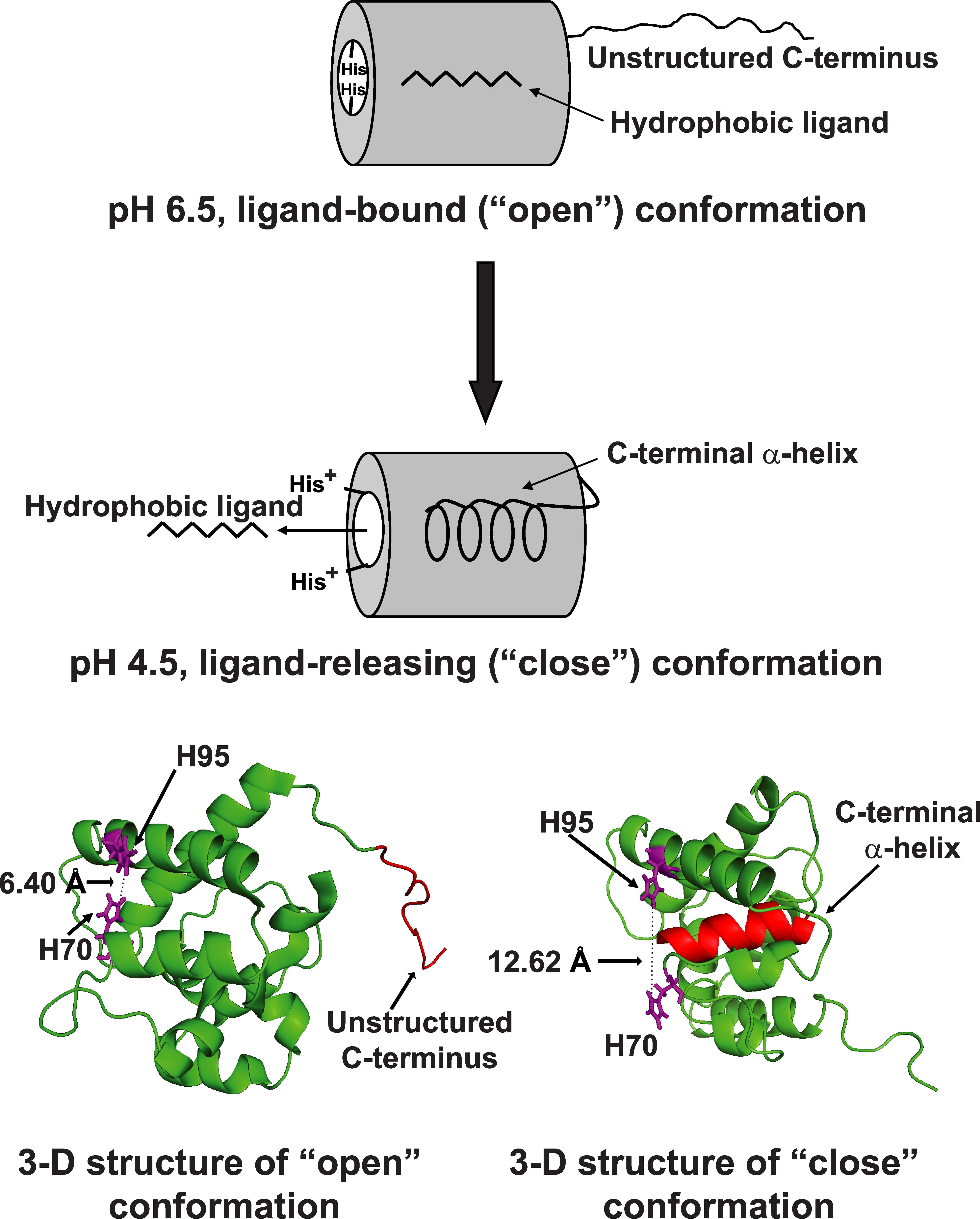
Ligand-bound ApolPBP1: a pH sensing protein (JBC, 2009)
Human PDZ domain and Tumorigenesis
Glutaminase-interacting protein (GIP) is a small 14 kDa protein containing a single PDZ domain. GIP was originally identified in a yeast two-hybrid genetic selection system in human brain while looking for interactors of glutaminase. GIPis directly involved in the modulation of tumor growth through regulation of glutaminase and b-catenin. In addition, we have found that GIP interacts with the cell surface protein FAS, which belongs to tumor necrosis factor (TNF) receptor family and mediates cell apoptosis. Apart from glutaminase, b-catenin and FAS, a plethora of binding partners has been reported implicating GIP in key biological processes.Indeed, all the signal transduction pathways involving GIP can lead to cancer when unregulated. Interestingly, GIP regulates many of these signaling processes through its PDZ domain. Because PDZ proteins have well-defined binding sites, they are promising targets for drug discovery. Furthermore, GIP is one of the smallest members of the PDZ family, containing only one PDZ domain that represents its full primary structure, thus offering it as a very suitable candidate for structural studies. Structure, function, and interaction studies of GIP with different binding partners will provide us the insight into the mechanisms of action of this multifunctional protein, which is indeed a necessary prelude for successful drug design.Our investigation into structure, function, kinetics, dynamics, and interaction studies of GIP with different binding partners will provide the insight into the mechanisms and role of this PDZ domain containing human protein playsin recognition, signaling and tumorigenesis, which is essential for successful drug design.
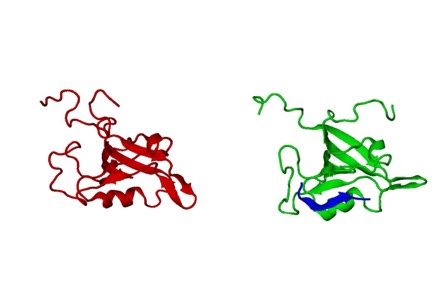
3D NMR Model of GIP and Its Complexes (Biochemistry, 2011 and 2012).
 Binding site residues mapping with acceptor peptide (Hung et. al., JBC, 2012).
Binding site residues mapping with acceptor peptide (Hung et. al., JBC, 2012).








